Details
| Basic Information |
| Taxanomy |
| Sequence Information |
| Modeled Structures |
| Gene Information |
| Gene Ontology |
| Family Classification |
Details : Q9FY94
dbSWEET id: dbswt_90
Accession: Q9FY94
Uniprot status: Reviewed
Organism: Arabidopsis thaliana
Kingdom: Plantae
Taxonomy back to top
Eukaryota ⇒ Viridiplantae ⇒ Streptophyta ⇒ Embryophyta ⇒ Tracheophyta ⇒ Spermatophyta ⇒ Magnoliophyta ⇒ eudicotyledons ⇒ Gunneridae ⇒ Pentapetalae ⇒ rosids ⇒ malvids ⇒ Brassicales ⇒ Brassicaceae ⇒ Camelineae ⇒ Arabidopsis.
Sequence Information back to top
Sequence length: 292
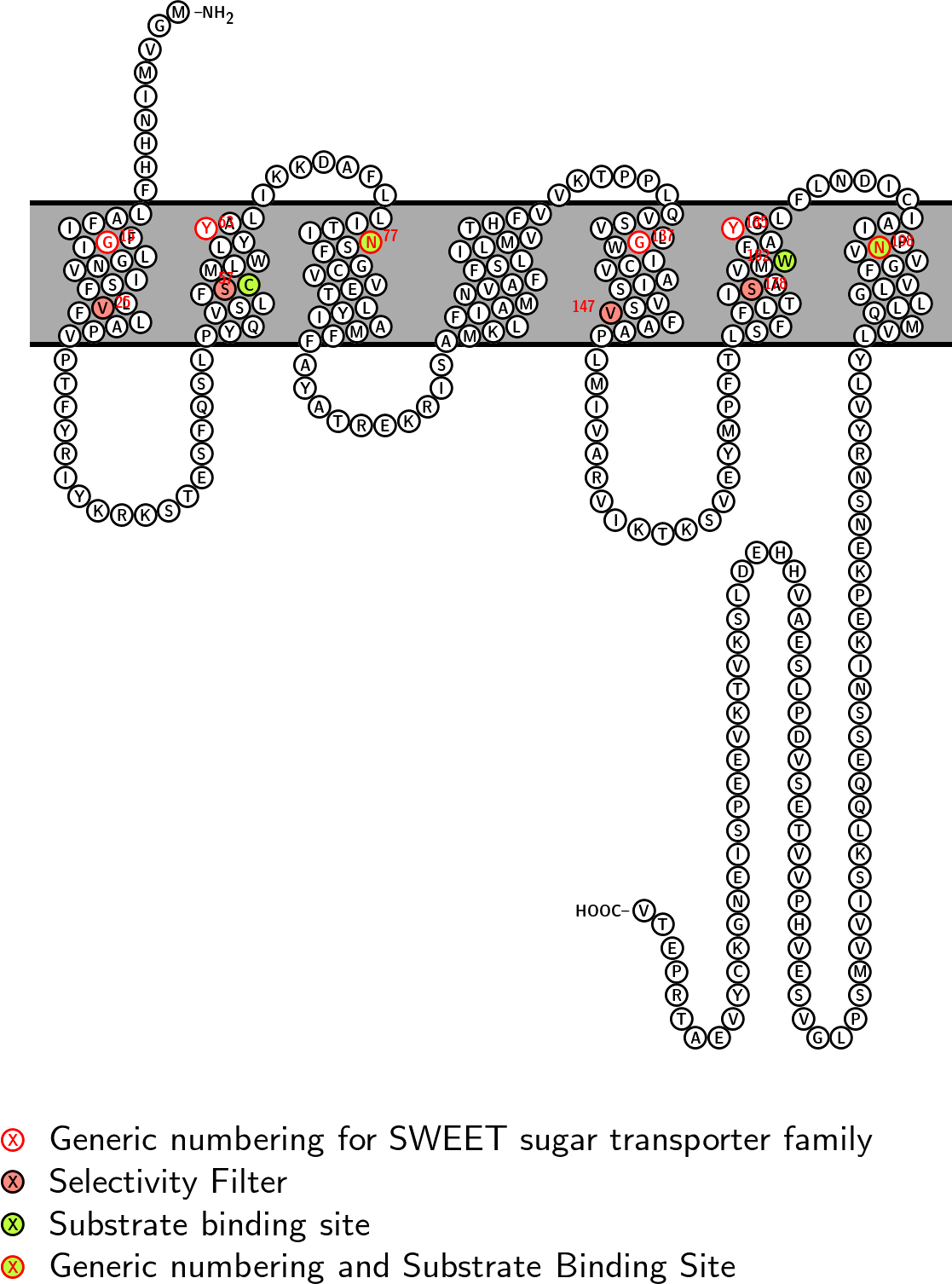
Substrate Binding Site: CNWN CVV: 441 CHI: -5.4
Selectivity Filter: VSVS CVV: 356 CHI: 6.8
Fasta sequence:
>sp|Q9FY94|SWT15_ARATH|Reviewed|Arabidopsis_thaliana|292
MGVMINHHFLAFIFGILGNVISFLVFLAPVPTFYRIYKRKSTESFQSLPYQVSLFSCMLW
LYYALIKKDAFLLITINSFGCVVETLYIAMFFAYATREKRISAMKLFIAMNVAFFSLILM
VTHFVVKTPPLQVSVLGWICVAISVSVFAAPLMIVARVIKTKSVEYMPFTLSFFLTISAV
MWFAYGLFLNDICIAIPNVVGFVLGLLQMVLYLVYRNSNEKPEKINSSEQQLKSIVVMSP
LGVSEVHPVVTESVDPLSEAVHHEDLSKVTKVEEPSIENGKCYVEATRPETV
Snake diagram with key positions labelled
Modeled Structuresback to top
Model start: 4 Model end: 217
Alignment file: Q9FY94.pir
Inward Open:
Template: 5CTG.pdb
Model structure: Q9FY94_inward.pdb
Procheck score ⇒ Ramachandran plot: 93.9% favored 5.6% allowed .5% week .0% disallowed
Outward Open:
Template: 5CTG_outward.pdb
Model structure: Q9FY94_outward.pdb
Procheck score ⇒ Ramachandran plot: 93.4% favored 4.6% allowed 1.5% week .5% disallowed
Occluded:
Template: 5CTG_occluded.pdb
Model structure: Q9FY94_occluded.pdb
Procheck score ⇒ Ramachandran plot: 92.3% favored 5.6% allowed 1.5% week .5% disallowed
Gene Informationback to top
Gene ID: 831156 Total Exons: 6 Coding Exons: 6
The diagram below shows the position of Exon-junctions in the protein structure ( Loops and transmembrane helices ).
If more than 1 exon translates a single secondary-structure , only the final exon is represented in figure.
Point on the red bars to view exon number







Gene Ontologyback to top
GO:0000139 - Golgi membrane
GO:0005887 - integral component of plasma membrane
GO:0008515 - sucrose transmembrane transporter activity
GO:0051119 - sugar transmembrane transporter activity
GO:0071215 - cellular response to abscisic acid stimulus
GO:0071470 - cellular response to osmotic stress
GO:0071446 - cellular response to salicylic acid stimulus
GO:0009793 - embryo development ending in seed dormancy
GO:0010150 - leaf senescence
GO:0010431 - seed maturation
GO:0015770 - sucrose transport
Family Classificationback to top
Pfam: PF03083: MtN3_slv.
Interpro: IPR004316: SWEET_sugar_transpr.
Panther: NA
