Details
| Basic Information |
| Taxanomy |
| Sequence Information |
| Modeled Structures |
| Gene Information |
| Gene Ontology |
| Family Classification |
Details : Q9CXK4
dbSWEET id: dbswt_985
Accession: Q9CXK4
Uniprot status: Reviewed
Organism: Mus musculus
Kingdom: Metazoa
Taxonomy back to top
Eukaryota ⇒ Metazoa ⇒ Chordata ⇒ Craniata ⇒ Vertebrata ⇒ Euteleostomi ⇒ Mammalia ⇒ Eutheria ⇒ Euarchontoglires ⇒ Glires ⇒ Rodentia ⇒ Sciurognathi ⇒ Muroidea ⇒ Muridae ⇒ Murinae ⇒ Mus ⇒ Mus.
Sequence Information back to top
Sequence length: 221
Substrate Binding Site: NNWN CVV: 451 CHI: -11.4
Selectivity Filter: MNMC CVV: 430 CHI: 2.8
Fasta sequence:
>sp|Q9CXK4|SWET1_MOUSE|Reviewed|Mus_musculus|221
MEAGGVADSFLSSACVLFTLGMFSTGLSDLRHMQRTRSVDNIQFLPFLTTDVNNLSWLSY
GVLKGDGTLIIVNSVGAVLQTLYILAYLHYSPQKHGVLLQTATLLAVLLLGYGYFWLLVP
DLEARLQQLGLFCSVFTISMYLSPLADLAKIVQTKSTQRLSFSLTIATLFCSASWSIYGF
RLRDPYITVPNLPGILTSLIRLGLFCKYPPEQDRKYRLLQT
Snake diagram with key positions labelled
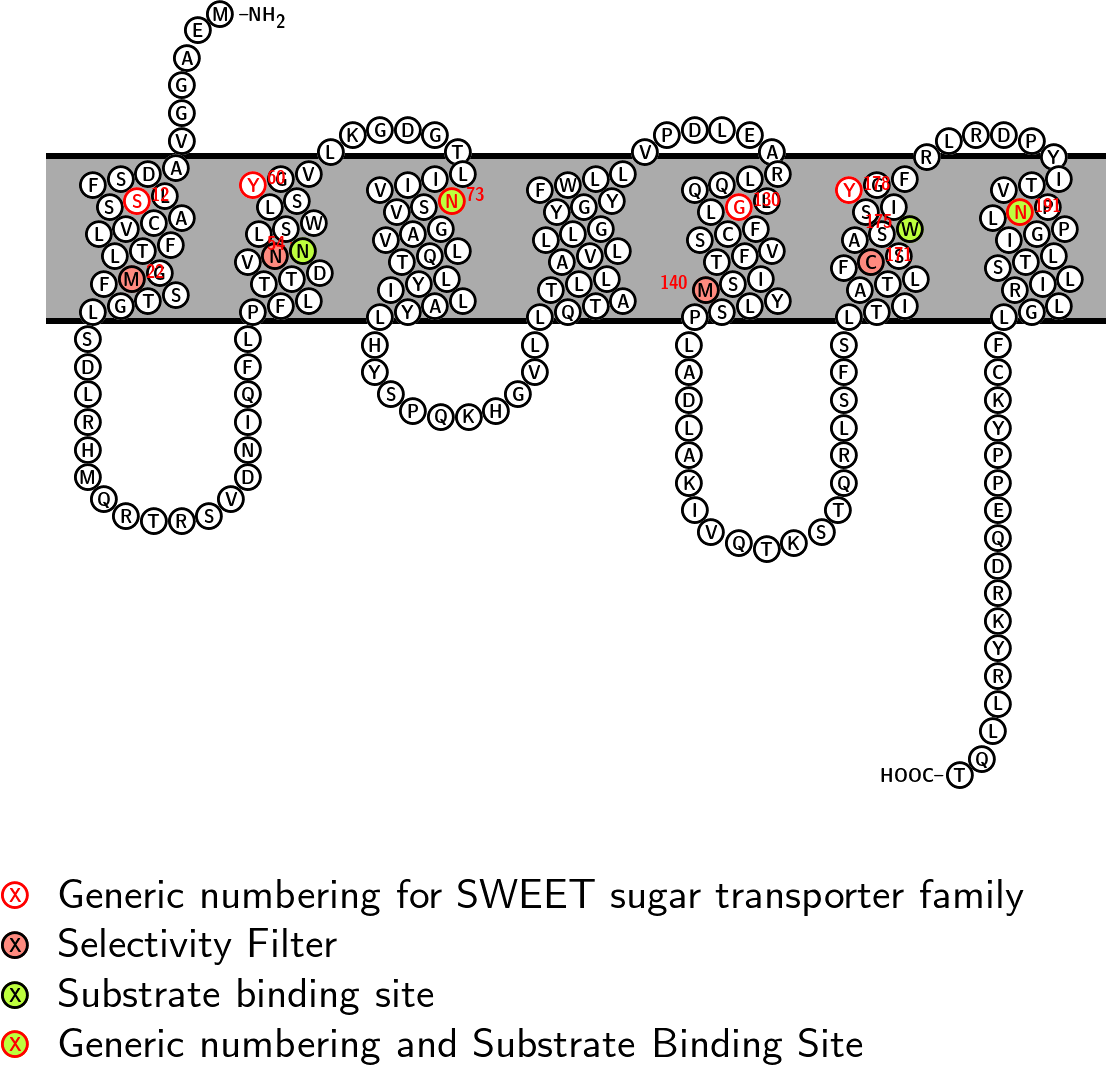
Modeled Structuresback to top
Model start: 1 Model end: 210
Alignment file: Q9CXK4.pir
Inward Open:
Template: 5CTG.pdb
Model structure: Q9CXK4_inward.pdb
Procheck score ⇒ Ramachandran plot: 90.3% favored 6.5% allowed 2.7% week .5% disallowed
Outward Open:
Template: 5CTG_outward.pdb
Model structure: Q9CXK4_outward.pdb
Procheck score ⇒ Ramachandran plot: 91.4% favored 7.0% allowed 1.6% week .0% disallowed
Occluded:
Template: 5CTG_occluded.pdb
Model structure: Q9CXK4_occluded.pdb
Procheck score ⇒ Ramachandran plot: 91.9% favored 6.5% allowed .5% week 1.1% disallowed
Gene Informationback to top
Gene ID: 19729 Total Exons: 6 Coding Exons: 6
The diagram below shows the position of Exon-junctions in the protein structure ( Loops and transmembrane helices ).
If more than 1 exon translates a single secondary-structure , only the final exon is represented in figure.
Point on the red bars to view exon number







Gene Ontologyback to top
GO:0005794 - Golgi apparatus
GO:0000139 - Golgi membrane
GO:0016021 - integral component of membrane
GO:0005634 - nucleus
GO:0005886 - plasma membrane
GO:0042947 - glucoside transmembrane transporter activity
GO:0051119 - sugar transmembrane transporter activity
GO:0008643 - carbohydrate transport
GO:0042946 - glucoside transport
GO:0045815 - positive regulation of gene expression epigenetic
Family Classificationback to top
Pfam: PF03083: MtN3_slv
Interpro: IPR004316: SWEET_sugar_transpr.
Panther: NA
