Details
| Basic Information |
| Taxanomy |
| Sequence Information |
| Modeled Structures |
| Gene Information |
| Gene Ontology |
| Family Classification |
Details : A0A101G2J5
dbSWEET id: dbswt_1535
Accession: A0A101G2J5
Uniprot status: Unreviewed
Organism: Actinobacteria bacterium
Kingdom: Bacteria
Sequence Information back to top
Sequence length: 139
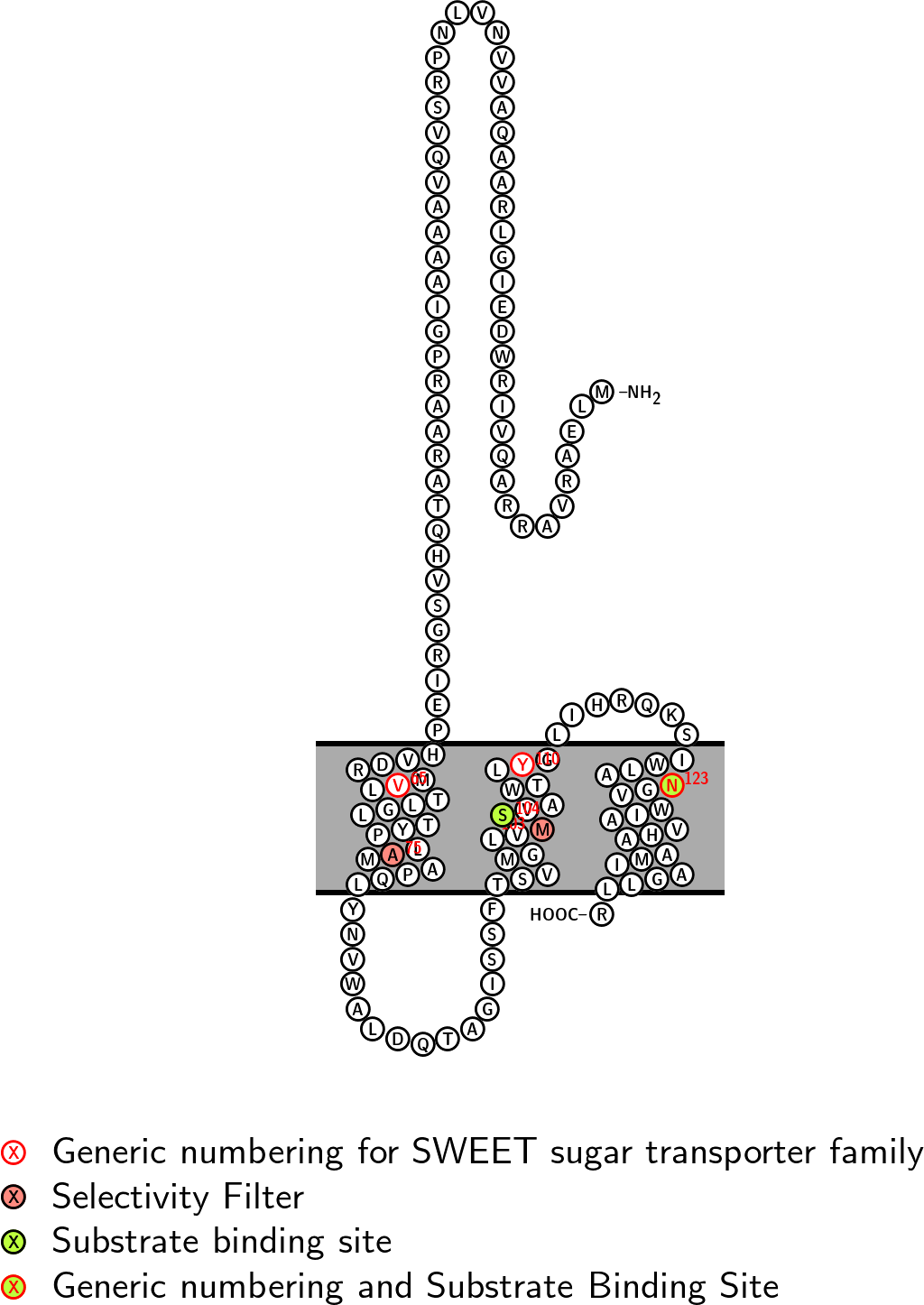
Substrate Binding Site: SNSN CVV: 338 CHI: -8.6
Selectivity Filter: AMAM CVV: 382 CHI: 7.4
Fasta sequence:
>tr|A0A101G2J5|A0A101G2J5_9ACTN|Unreviewed|Actinobacteria bacterium| 139
MLEARVARRAQVIRWDEIGLRAAQAVVNVLNPRSVQVAAAAIGPRAARATQHVSGRIEPH
VDRMVLTLGLTYPLAMAPQLYNVWALDQTAGISSFTSVMGLVMSVAWTLYGLIHRQKSIW
LANGVWIAVHAAMIAGLLR
Snake diagram with key positions labelled
Modeled Structuresback to top
Model start: 63 Model end: 139 Inward Open: Template: 4X5M.pdb Model structure: A0A101G2J5_inward.pdb Alignment file: A0A101G2J5_inw.pir Procheck score ⇒ Ramachandran plot: 90.9% favored 9.1% allowed .0% week .0% disallowed Outward Open: Template: 4X5N.pdb Model structure: A0A101G2J5_outward.pdb Alignment file: A0A101G2J5_out.pir Procheck score ⇒ Ramachandran plot: 94.7% favored 5.3% allowed .0% week .0% disallowed Occluded: Model structure: A0A101G2J5_occluded.pdb Alignment file: A0A101G2J5_occ.pir Procheck score ⇒ Ramachandran plot: 93.2% favored 6.1% allowed .0% week .8% disallowed
Family Classificationback to top
Pfam: PF03083: MtN3_slv
Interpro: IPR004316: SWEET_sugar_transpr.
Panther: NA
